Cunningham, Lucas, Lingley, Jessica, Tirados, Inaki  ORCID: https://orcid.org/0000-0002-9771-4880, Esterhuizen, Johan, Opiyo, Mercy, Mangwiro, Clement, Lehane, Mike and Torr, Steve
ORCID: https://orcid.org/0000-0002-9771-4880, Esterhuizen, Johan, Opiyo, Mercy, Mangwiro, Clement, Lehane, Mike and Torr, Steve  ORCID: https://orcid.org/0000-0001-9550-4030
(2020)
'Evidence of the absence of Human African Trypanosomiasis in two Northern Districts of Uganda: analyses of cattle, pigs and tsetse flies for the presence of Trypanosoma brucei gambiense'. PLoS Neglected Tropical Diseases, Vol 14, Issue 4, e0007737.
ORCID: https://orcid.org/0000-0001-9550-4030
(2020)
'Evidence of the absence of Human African Trypanosomiasis in two Northern Districts of Uganda: analyses of cattle, pigs and tsetse flies for the presence of Trypanosoma brucei gambiense'. PLoS Neglected Tropical Diseases, Vol 14, Issue 4, e0007737.
|
Text
PNTD-D-19-01371R1.docx - Accepted Version Available under License Creative Commons Attribution. Download (1MB) |
||
![[img]](https://archive.lstmed.ac.uk/14102/2.hassmallThumbnailVersion/image5.png)
|
Image
image5.png - Supplemental Material Available under License Creative Commons Attribution. Download (393kB) | Preview |
|
![[img]](https://archive.lstmed.ac.uk/14102/3.hassmallThumbnailVersion/image4.png)
|
Image
image4.png - Supplemental Material Available under License Creative Commons Attribution. Download (209kB) | Preview |
|
![[img]](https://archive.lstmed.ac.uk/14102/4.hassmallThumbnailVersion/image3.png)
|
Image
image3.png - Supplemental Material Available under License Creative Commons Attribution. Download (132kB) | Preview |
|
![[img]](https://archive.lstmed.ac.uk/14102/5.hassmallThumbnailVersion/image2.png)
|
Image
image2.png - Supplemental Material Available under License Creative Commons Attribution. Download (47kB) | Preview |
|
![[img]](https://archive.lstmed.ac.uk/14102/6.hassmallThumbnailVersion/image1.png)
|
Image
image1.png - Supplemental Material Available under License Creative Commons Attribution. Download (807kB) | Preview |
Abstract
Background
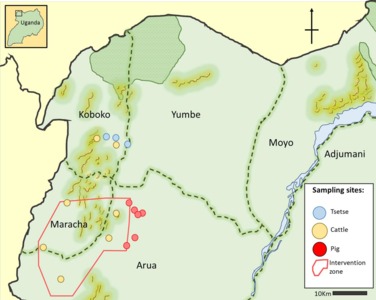
Large-scale control of sleeping sickness has led to a decline in the number of cases of Gambian human African trypanosomiasis (g-HAT) to <2000/year. However, achieving complete and lasting interruption of transmission may be difficult because animals may act as reservoir hosts for T. b. gambiense. Our study aims to update our understanding of T. b. gambiense in local vectors and domestic animals of N.W. Uganda.
Methods
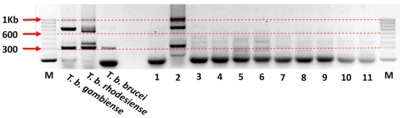
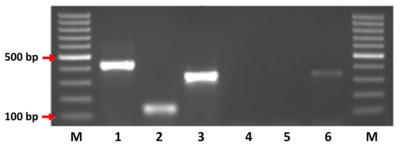
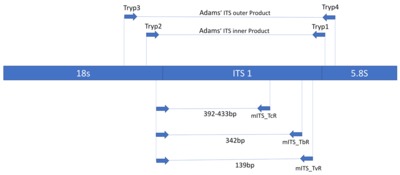
We collected blood from 2896 cattle and 400 pigs and In addition, 6664 tsetse underwent microscopical examination for the presence of trypanosomes. Trypanosoma species were identified in tsetse from a subsample of 2184 using PCR. Primers specific for T. brucei s.l. and for T. brucei sub-species were used to screen cattle, pig and tsetse samples.
Results
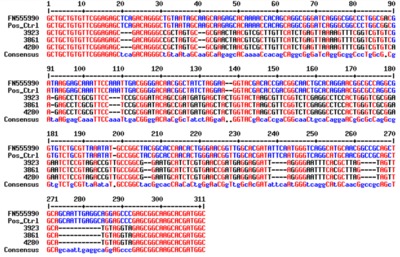
In total, 39/2,088 (1.9%; 95% CI=1.9-2.5) cattle, 25/400 (6.3%; 95% CI=4.1-9.1) pigs and 40/2,184 (1.8%; 95% CI=1.3-2.5) tsetse, were positive for T. brucei s.l.. Of these samples 24 cattle (61.5%), 15 pig (60%) and 25 tsetse (62.5%) samples had sufficient DNA to be screened using the T. brucei sub-species PCR. Further analysis found no cattle or pigs positive for T. b. gambiense, however, 17/40 of the tsetse samples produced a band suggestive of T. b. gambiense. When three of these 17 PCR products were sequenced the sequences were markedly different to T. b. gambiense, indicating that these flies were not infected with T. b. gambiense.
Conclusion
The lack of T. b. gambiense positives in cattle, pigs and tsetse accords with the low prevalence of g-HAT in the human population. We found no evidence that livestock are acting as reservoir hosts. However, this study highlights the limitations of current methods of detecting and identifying T. b. gambiense which relies on a single copy-gene to discriminate between the different sub-species of T. brucei s.l.
| Item Type: | Article |
|---|---|
| Subjects: | QW Microbiology and Immunology > Viruses > QW 162 Insect viruses QX Parasitology > QX 4 General works QX Parasitology > Insects. Other Parasites > QX 500 Insects WA Public Health > Health Problems of Special Population Groups > WA 395 Health in developing countries WC Communicable Diseases > Tropical and Parasitic Diseases > WC 705 Trypanosomiasis |
| Faculty: Department: | Biological Sciences > Vector Biology Department |
| Digital Object Identifer (DOI): | https://doi.org/10.1371/journal.pntd.0007737 |
| Depositing User: | Samantha Sheldrake |
| Date Deposited: | 08 Apr 2020 12:05 |
| Last Modified: | 09 Apr 2020 10:08 |
| URI: | https://archive.lstmed.ac.uk/id/eprint/14102 |
Statistics
Actions (login required)
 |
Edit Item |

 Tools
Tools Tools
Tools